Planned Research A01: Functional analysis of neo-self
▼Mechanisms for the creation of neo-self in the thymus ▼Generation mechanisms of neo-self by metal or medicine
▼Generation of neo-self antigens in the tumor microenvironment ▼Repertoire analysis of neo-self-recognizing receptors
Mechanisms for the creation of neo-self in the thymus
| Principal Investigator |  |
Mitsuru Matsumoto | Professor, Institute for Enzyme Research, Tokushima University Lab Website (http://www.tokushima-u.ac.jp/ier/english/research/immunity.html) |
| Co-Investigator (buntan) |  |
Yasunobu Yoshikai | Academic Researcher, Professor Emeritus, Division of Host Defense, Medical Institute of Bioregulation, Kyushu University Lab Website (http://www.bioreg.kyushu-u.ac.jp/kansenseigyo/english-home0.html) |
| Co-Investigator (renkei) | Shigeo Murata | Professor, Laboratory of Protein Metabolism, Graduate School of Pharmaceutical Science, The University of Tokyo |
Matsumoto Group
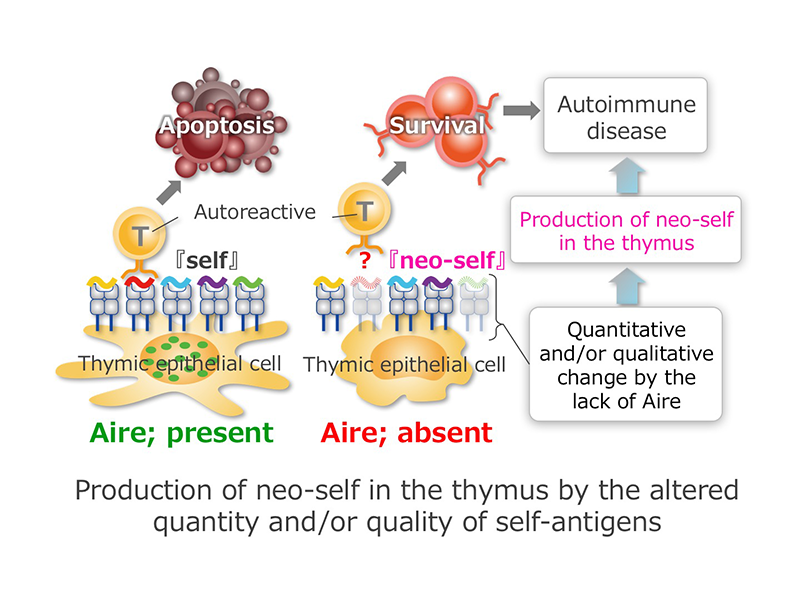
Thymus is a major organ that plays a central role in the elimination of autoreactive T cells, and thymic epithelial cells (TECs) have a unique ability to express wide varieties of immunological self to achieve this task. Aire, a transcription factor whose bona fide target genes have yet to be determined, contributes to this process. Accordingly, human and mice lacking the normal function of Aire develop organ-specific autoimmune diseases. However, the exact function of Aire in establishing immunological tolerance remains unknown. In this research project, we will investigate how the lack of Aire in TECs results in the breakdown of self-tolerance. Especially, we will try to test whether Aire is contributing to the establishment of self-tolerance simply by regulating the expression levels of immunological self. In this regard, we have a working hypothesis that loss of Aire in TECs will create a “neo-self” in which establishment of self-tolerance is perturbed by the mechanism that needs further investigation.
J Immunol., 195(11):5149-58, 2015. doi: 10.4049/jimmunol.1501000.
J Immunol., 195(10):4641-9, 2015. doi: 10.4049/jimmunol.1501026.
J Immunol., 192(6):2585-92, 2014. doi: 10.4049/jimmunol.1302786.
Eur J Immunol., 41(1):12-7, 2011. doi: 10.1002/eji.201041024.
J Exp Med., 207(5):963-71, 2010. doi: 10.1084/jem.20092144.
Yoshikai Group
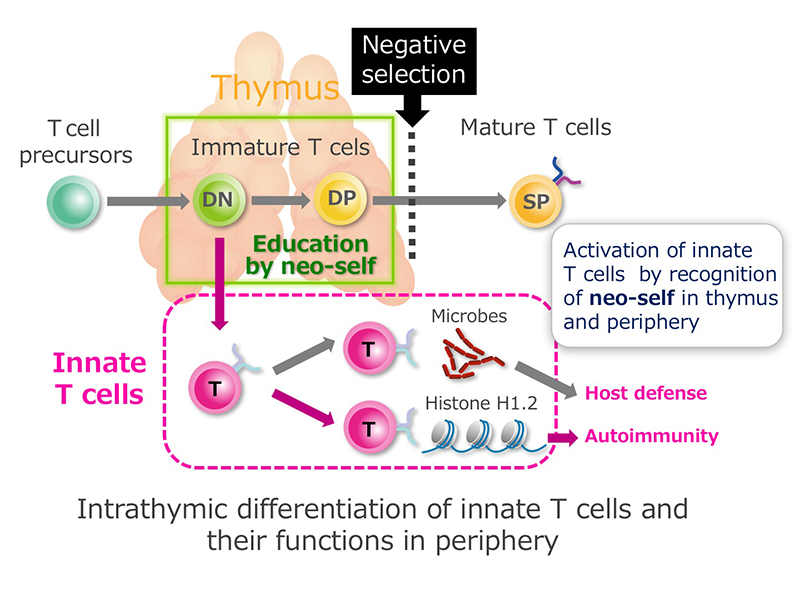
Conventional T cells, which are exported from the thymus after negative selection as naïve cells and acquire effector functions upon antigen encounter in the periphery. On the other hand, innate T cells are functionally committed into effector cells showing abilities of cytokine production and cytotoxicity within thymus and are disproportionately distributed in mucosal epithelia. Preceding the generation of conventional T cells from double-positive (DP) cells as naive cells, early T cell precursors at the double-negative (DN) stages are committed to innate T cells in fetal thymus. In the present study, we investigate “neo-self”, which innate T cells recognize for differentiation in thymus and for host defense and autoimmunity in periphery. The discovery of “neo-self” recognized by innate T cells may challenge the conventional immunological paradigm to be sub-classified into innate and adaptive immunity.
Nat Commun., 25;6:7464, 2015. doi: 10.1038/ncomms8464.
J Autoimmun., 57:14-23, 2015. doi: 10.1016/j.jaut.2014.11.005.
J Immunol., 192(5):2210-8, 2014. doi: 10.4049/jimmunol.1302145.
Mucosal Immunol., 6(6):1191-201, 2013. doi: 10.1038/mi.2013.18.
Blood., 118(3):586-93, 2011. doi: 10.1182/blood-2011-02-334995.
Generation mechanisms of neo-self by metal or medicine
| Principal Investigator |  |
Kouetsu Ogasawara | Professor, Department of Immunobiology, Institute of Development, Aging and Cancer, Tohoku University Lab Website (http://www2.idac.tohoku.ac.jp/dep/imbio/) |
Ogasawara Group
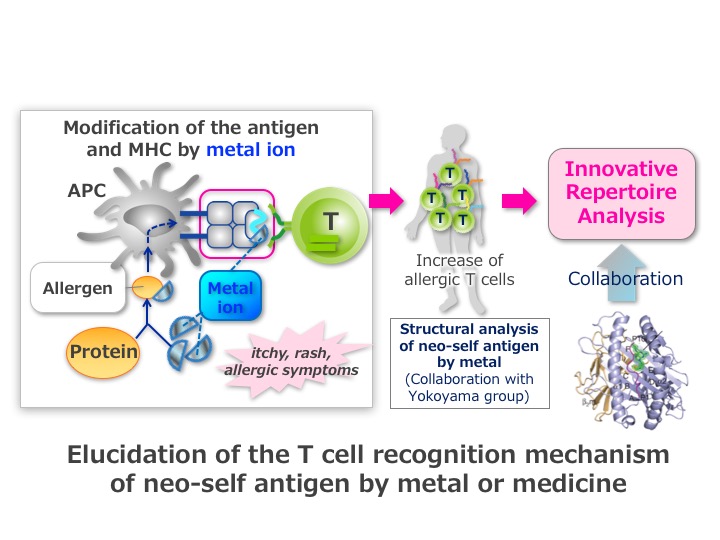
In the immune response, protein is decomposed and characteristic peptide is presented as an antigen. So far, many analysis of the T cell receptor (TCR) to the peptide have been performed. However, since metal or medicine exist as an ion or a low molecular compound in vivo, it has not been understood how these molecules are presented as an antigen. Metal or medicine are called hapten, and hapten by itself does not have the antigenicity. However, hapten exerts immunological effects by modifying the peptide of self (self), which are considered to constitute new self-antigens (neo-self).
Because T cells are able to respond to the huge variety of pathogenic microbes, TCRs have large repertoires. To determine the specific T-cell to respond to the antigen, it is necessary to comprehensively analyze TCR repertoires. We therefore have developed a system to comprehensively measure TCR repertoires. Using this new technology, we investigate how “neo-self” is generated by metal or medicine.
Int Immunopharmacol., 35:70-6, 2016. doi: 10.1016/j.intimp.2016.03.021.
Exp Hematol., 44(2):116-24, 2016. doi: 10.1016/j.exphem.2015.11.002.
PLoS One., 9(2):e86810, 2014. doi: 10.1371/journal.pone.0086810.
Proc Natl Acad Sci U S A., 110(23):9421-6, 2013. doi: 10.1073/pnas.1300140110
Proc Natl Acad Sci U S A., 108(45):18360-5, 2011. doi: 10.1073/pnas.1110584108.
Generation of neo-self antigens in the tumor microenvironment
| Principal Investigator |  |
Keiko Udaka | Professor, Department of Immunology, School of Medicine, Kochi University University Website (http://www.kochi-ms.ac.jp/kms_e/) Lab Website (http://www.kochi-ms.ac.jp/~ff_immnl/) |
| Co-Investigator (buntan) |  |
Yasuharu Nishimura | Academic Researcher, Professor Emeritus, Department of Immunology, Graduate School of Medical Sciences, Kumamoto University Lab Website (https://www.immunology-kumamoto.com/) |
| Co-Investigator (renkei) | Hiroyoshi Nishikawa | Director, Division of cancer Immunology, Exploratory Oncology Research and Clinical Trial Center, Research Institute, National Cancer Center Professor, Department of Immunology, Nagoya University Graduate School of Medicine |
Udaka Group

The recent introduction of immune-checkpoint inhibitors has changed the treatment of cancer patients dramatically. Further boosting of the targeted T cell responses is pivotal to improve the efficacy of immunotherapy as well as to reduce autoimmune responses against normal tissues. Our team investigate the antigen presentation taking place in the tumor microenvironment and delineate the mechanism how tumor cells, a component of self, are recognized as “neo-self” by T cells. Recently, we found that tumor endothelial cells (ECs) have the capacity to present tumor antigens. Such ECs serve as a gate to invite tumor-specific Th cells and induce subsequent infiltration of CTLs. By exploiting these knowledge we hope to develop a method to induce tumor antigen-specific CTLs and Th cells and control established tumors. A computational method to identify MHC class I- and class II-binding peptides we have developed should help identify target peptides and design proper immunization protocols in combination with adjuvants. At last, we challenge to develop a technology to control the peptide-specific immune responses positively or negatively.
Eur J Immunol., 39(1):96-112, 2009. doi: 10.1002/eji.200838796.
J Immunol., 169(10): 5744-53, 2002.
J Immunol., 164(4):1873-80, 2000.
J Immunol., 157(2): 670-8, 1996.
Cell., 69(6):989-98, 1992.
Nishimura Group
T cells recognize tumor-associated neo-self antigenic peptides in the context of HLA molecules expressed on the surface of tumor cells to recognize and eliminate tumor cells. HLA genes are the most polymorphic genes among the human genomes and miss-match of HLA alleles induces a strong rejection of a transplanted allograft. This polymorphism of HLA molecules is focused on amino acid residues consisting of the peptide-binding groove of the HLA molecule. As a result, the amino acids sequences of the peptides bound by allelic products of HLA genes are different. HLA molecules are divided into two classes, namely HLA class I expressed on all nucleated cells and HLA class II mainly expressed on antigen-presenting cells such as dendritic cells. HLA class I molecules present tumor-associated neo-self antigenic short peptides usually consisting of 9~12 amino acids on the surface of tumor cells, and CD8+ cytotoxic T lymphocyte (CTL) recognizes these complexes to kill tumor cells. On the other hand, HLA class II molecules present tumor-associated neo-self antigenic long peptides usually consisting of 12~26 amino acids on the surface of antigen-presenting cells, and CD4+ helper T (Th) cells recognize these complexes to activate other immune cells by producing various kinds of cytokines.
In this research project, I will identify new tumor-associated neo-self antigenic long peptides that can activate not only HLA class II-restricted Th cells but also HLA class I-restricted CTL to induce more potent tumor immunity. Moreover I will investigate the T-cell receptor (TCR) gene usages of tumor-reactive T cells induced by the peptides, and investigate whether TCR-gene sequencing of peripheral T cells isolated from cancer patients immunized with peptides vaccine is useful for immune monitoring of the patients or not. I will also investigate mechanisms of immune suppression of T cells in tumor-bearing hosts, and develop a new therapy for cancer patients by liberation from immunosuppressive status. The possibility of combined cancer immunotherapy described above will be also investigated.
Nat Commun., 6:6702, 2015. doi: 10.1038/ncomms7702.
Clin Cancer Res., 21(2):312-21, 2015. doi: 10.1158/1078-0432.CCR-14-0202.
Onco Immunology., 3:e28100, 2014. doi:10.4161/onci.28100.
Science., 345(6195):455-9, 2014. doi: 10.1126/science.1249749.
Clin Cancer Res., 19(16):4508-20, 2013. doi: 10.1158/1078-0432.CCR-13-0197.
Repertoire analysis of neo-self-recognizing receptors
| Principal Investigator |  |
Hiroyuki Kishi | Associate Professor, Department of Immunology, Graduate School of Medicine and Pharmaceutical Sciences (Medicine), University of Toyama Lab Website (http://www.med.u-toyama.ac.jp/immuno/top.html) |
| Co-Investigator (renkei) | Hiroshi Hamana | Assistant Professor, Department of Immunology, Graduate School of Medicine and Pharmaceutical Sciences (Medicine), University of Toyama | |
| Tatsuhiko Ozawa | Assistant Professor, Department of Immunology, Graduate School of Medicine and Pharmaceutical Sciences (Medicine), University of Toyama |
Kishi Group
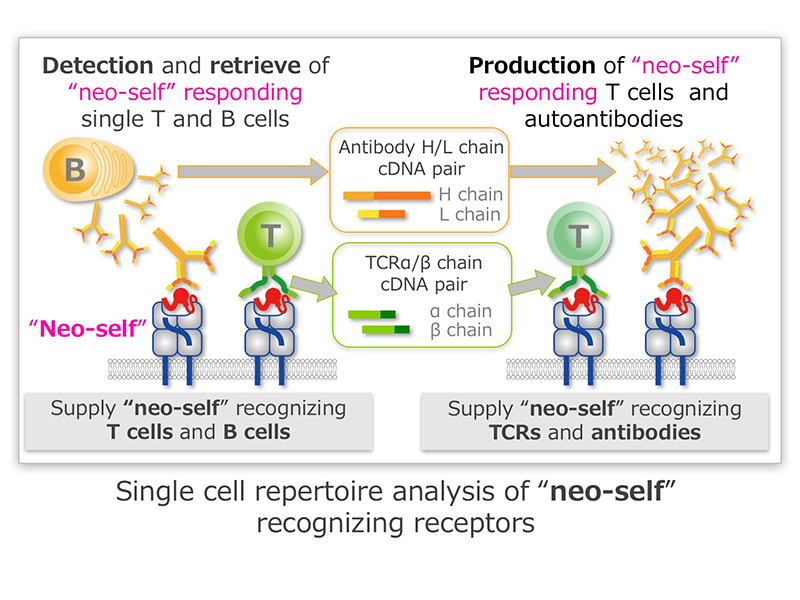
In the “neo-self” Research Area, we clarify the mechanisms of the development of autoimmune diseases or allergy and the immune regulation of tumor by analyzing “neo-self”, and develop the methods to regulate these diseases. Antigen receptors play important roles in recognition of “neo-self” by immune cells. Our research group first develop the simple method to detect “neo-self”-responding T cells or B cells to obtain “neo-Self”-recognizing receptors from the single cells. We next analyze the interaction of “neo-self” and the receptors at cellular or molecular levels in detail. Specifically, by using the “neo-self” that is identified by other research groups in the Area, we prepare materials for detecting the “neo-self”, such as “neo-self” multimers or “neo-self”-presenting cells, use them to identify the “neo-self”-recognizing cells and clone their T cell receptors or B cell receptors rapidly and with high efficiency. We then analyze the reaction of the receptors to the “neo-self” at cellular and molecular levels, by analyzing the response of immune cells that express the “neo-self” receptors or analyzing the interaction of the soluble “neo-self” and their soluble receptors with X-ray crystallography.
Sci Transl Med., 7(310):310ra167, 2015. doi: 10.1126/scitranslmed.aac5477.
Arthritis Rheumatol., 67(8):2020-31, 2015. doi: 10.1002/art.39161.
Biochem Biophys Res Commun., 453(4):798-803, 2014. doi: 10.1016/j.bbrc.2014.10.024.
Nat Med., 19(11):1542-6, 2013. doi: 10.1038/nm.3358.
Nat Protoc., 6(5):668-76, 2011. doi: 10.1038/nprot.2011.322.
