Planned Research A02: Structural analysis of neo-self
▼Structural analysis of neo-self ▼Analysis for neo-self in MHC-misfolded protein complex ▼Genetic analysis of neo-self
Structural analysis of neo-self
| Principal Investigator |  |
Shigeyuki Yokoyama | Distinguished Senior Scientist, RIKEN Structural Biology Laboratory RIKEN Website (http://www.riken.jp/en/research/labs/distinguished/struct_biol/) Lab Website (http://sbl.riken.jp/) |
| Co-Investigator (buntan) |  |
Takehiko Sasazuki | University Professor, Kyushu University Institute for Advanced Study University Website (http://ias.kyushu-u.ac.jp/eng/organization/sasazuki_01.php) |
| Co-Investigator (renkei) | Ken Yamamoto | Professor, Department of Medical Biochemistry, Kurume University School of Medicine |
Yokoyama Group
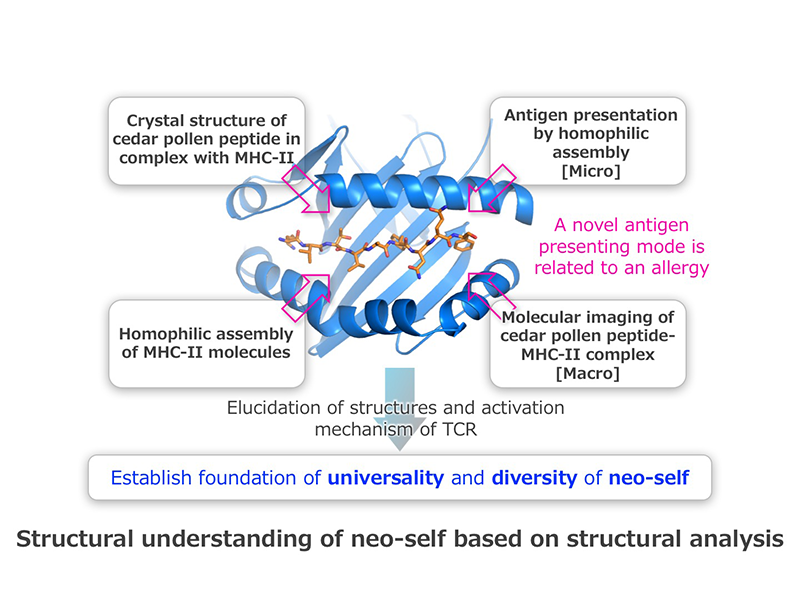
Human major histocompatibility complex (HLA) molecules specifically bind to antigenic peptides, from either self or non-self, and then the HLA–peptide complexes are recognized by T-cell receptors (TCRs). We determined the crystal structure of HLA-DP5, an HLA class II (HLA-II) molecule, in complex with cedar-pollen peptide. In collaboration with Dr. Sasazuki, Co-Investigator, we discovered that the HLA-DP5 molecules can interact with each other in vitro and in vivo. In this study, we compare and verify the antigen-presenting modes using the “neo-self” structural information of HLA-II molecules in complex with allergy antigens, self-antigens of autoimmune diseases, and mutant antigens of cancer immunity. Furthermore, we compare and verify the “neo-self”-TCR binding modes, based on the structural information of the complexes, and thereby systematize the relationships between the antigen-presenting mode and the immune response. From these analyses, we will identify the decisive characteristics that distinguish between neo-self and self, in efforts to contribute to the elucidation of the pathogenic mechanisms of immune diseases and to the development of new cures.
Nature., 531(7592):122-5, 2016. doi: 10.1038/nature16991.
Nature., 520(7547):312-6, 2015. doi: 10.1038/nature14301.
J Mol Biol., 426(17):3016-27, 2014. doi: 10.1016/j.jmb.2014.06.020.
Nature., 510(7506):507-11, 2014. doi: 10.1038/nature13440.
Science., 340(6128):75-8, 2013. doi: 10.1126/science.1229521.
Sasazuki Group
HLA shows extremely high polymorphism and binds foreign or self peptides to induce T cell activation which exerts protection of infectious diseases as well as development of allergy and autoimmune diseases. HLA-DP5 binds peptide (Crij-1) from Japanese cedar pollens to activate Crij-1 specific T cells which help B cells to produce IgE antibody to cedar pollens. This is the start of cedar pollinosis (typeⅠallergy). HLA-DP5 also binds peptide from thyroid stimulating hormone receptor (TSHR) to activate TSHR specific T cells which help B cells to produce agonistic IgG autoantibody to TSHR. This is the start of autoimmune thyroid disease (Greaves disease). In order to clarify the structural basis of HLA - peptide - T cell receptor (TCR) complex involved in allergic response or autoimmune response, we will analyze their crystal structures of same HLA-DP5 molecules with different allergic Crij-1 peptides or self TSHR peptides together with specific TCRs. Using the same strategy, we will also analyze crystal structures of HLA - peptide - TCR involved in immune regulation (Treg), and graft versus host disease (GvHD). We also try to identify the specific HLA molecules and their binding peptides from HLA-A*33:03-C*14:03-B*44:03-DRB1*13:02-DQB1*06:04-DPB1*04:01 haplotype which is involved in strong down regulation of immune response in the development of Graves disease and Hashimoto thyroiditis (Collaboration with S. Yokoyama).
Blood., 125(7):1189-97, 2015. doi: 10.1182/blood-2014-10-604785.
J Clin Endocrinol Metab., 100(2):E319-24, 2015. doi: 10.1210/jc.2014-3431.
J Mol Biol., 426(17):3016-27, 2014. doi: 10.1016/j.jmb.2014.06.020.
J Clin Endocrinol Metab., 99(2):E379-83, 2014. doi: 10.1210/jc.2013-2841.
Blood., 115(23):4664-70, 2010. doi: 10.1182/blood-2009-10-251157.
N Engl J Med., 339(17):1177-85, 1998.
Analysis for neo-self in MHC-misfolded protein complex
| Principal Investigator |  |
Tadashi Yokosuka | Professor, Department of Immunology, Tokyo Medical University University Website (http://www.tokyo-med.ac.jp/faculty/med/course/course18.html) Lab Website (http://tokyo-med-imm.jimdo.com/) |
| Co-Investigator (buntan) |  |
Tadahiro Suenaga | Associate Professor, Fukushima Medical University |
| Co-Investigator (renkei) | Hisashi Arase | Professor, Department of Immunochemistry, Research Institute for Microbial Diseases, Osaka University |
Yokosuka Group
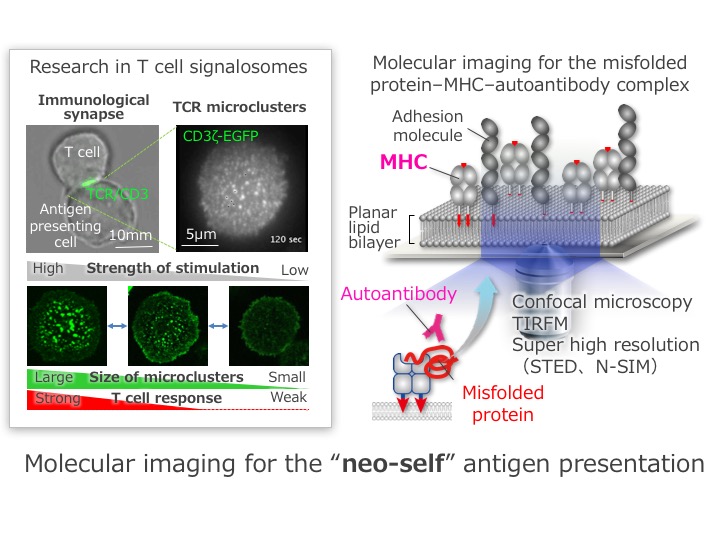
T cells contact with the other cells to know whether those cells are “self” or “non-self”. Otherwise they tightly conjugate with antigen presentence cells (APCs) to learn what are presented on the cell surface of APCs. Such a T cell–APC interface, “immunological synapse”, is constructed by a signalosome, “TCR microcluster”, which is an assembly of a few decades of T cell receptors (TCRs) gathering together with peptide-MHCs. We discovered this characteristic structure as a minimal unit for T cell activation by using an epoch-making strategy, the combination of a planar lipid bilayer system with single molecule imaging. Those imaging data consequently suggest us that a T cell recognizes “self” or “non-self” through TCR microclusters. In this research group, we are performing a high-tech imaging analysis on the basis of the former biochemical and biophysical studies. We will finally unveil the molecular interaction of an MHC–misfolding protein complex by a spatiotemporal approach to reveal the mechanisms of the immune tolerance disruption and to make a scientific foundation of molecular biology to overcome diseases and to develop a diagnostic strategy.
J Exp Med., 213(8):1609-25, 2016. doi: 10.1084/jem.20151088.
Nat Commun., 6:5555, 2015. doi: 10.1038/ncomms6555.
Nat Immunol., 15(5):465-72, 2014. doi: 10.1038/ni.2866.
Nat Immunol., 14(8):858-66, 2013. doi: 10.1038/ni.2634.
J Exp Med., 209(6):1201-17, 2012. doi: 10.1084/jem.20112741.
Suenaga Group

Autoimmune diseases are caused by immune response against self-antigens. However, it has remained unclear how immune response against self-antigens is induced. Misfolded proteins produced intracellularly are degraded and are not transported outside the cells. We have recently found that misfolded proteins associated with MHC class II molecules in ER escape from the degradation and are transported to the cell surface by aberrantly expressed MHC class II molecules (Figure). Because structure of misfolded proteins associated with MHC class II molecules are different from normal antigen, they seem to have different antigenicity from normal antigens. We propose that misfolded proteins associated with MHC class II molecules may initiate abnormal immune response and thus are involved in autoimmune diseases as neo-self antigen. Indeed, we have demonstrated that misfolded proteins associated with MHC class II molecules are major targets for autoantibodies produced in patients with autoimmune diseases such as rheumatoid arthritis and anti-phospholipid antibody syndrome. Furthermore, the autoantibody binding to neo-self complex is strongly associated with autoimmune disease susceptibility conferred by each MHC-II allele. These results suggest that neo-self complex generated by MHC class II molecules is a cause of autoimmune diseases. We will further analyze the mechanism of autoimmune diseases caused by neo-self complex from several points of view.
Adv Immunol., 129:1-23, 2016. doi: 10.1016/bs.ai.2015.09.005.
Blood., 125(18):2835-44, 2015. doi: 10.1182/blood-2014-08-593624
J Biochem., 158(5):367-72, 2015. doi: 10.1093/jb/mvv093.
Proc Natl Acad Sci USA., 111(10):3787-92, 2014. doi: 10.1073/pnas.1401105111.
Int Immunol., 25(4):235-46, 2013. doi: 10.1093/intimm/dxs155.
Genetic analysis of neo-self
| Principal Investigator |  |
Takashi Shiina | Professor, Department of Molecular Life Science, Division of Basic Medical Science and Molecular Medicine, Tokai University School of Medicine Lab Website (http://mls.med.u-tokai.ac.jp/mlsEngTop.html) |
| Co-Investigator (buntan) |  |
Kazuyoshi Hosomichi | Associate Professor, Department of Bioinformatics and Genomics, Graduate School of Advanced Preventive Medical Sciences, Kanazawa University Lab Website (http://big.w3.kanazawa-u.ac.jp/) |
| Co-Investigator (renkei) | Akira Oka | Lecturer, The Institute of Medical Science, Tokai University |
Shiina Group

The Human Leukocyte Antigen (HLA) genomic region has been related with more than 100 different diseases, including common chronic diseases and various autoimmune disorders. However, the simple question "What specific HLA alleles are genetically related with the diseases" has not been answered in most cases so far. No previous studies have focused specifically on HLA and other gene expression and polymorphisms that are involved in the generation of neo-self. Comprehensive genetic studies using innovative NGS technologies are necessary to elucidate the molecular mechanisms involved in the generation of neo-self.
The aim of this project is to elucidate genetic factors of the generation of neo-self by using the following three approaches: (1) genomics to detect variations in whole HLA gene regions from promoter-enhancer region to 3’ untranslated region and in neo-self related genes (e.g. autoantigen genes) associated with HLA-related diseases, (2) transcriptomics to detect RNA expression levels among HLA and neo-self related gene’s alleles, and identify novel splice variants and small RNAs, and (3) epigenetics to detect DNA methylation sites, histone modification sites and chromatin structures that affect HLA and neo-self related gene expression levels.
Immunology. 2016 Jul 10. doi: 10.1111/imm.12624.
Haematologica., 101(4):491-8, 2016. doi: 10.3324/haematol.2015.136903.
Nat Commun., ;7:10356, 2016. doi: 10.1038/ncomms10356.
BMC Genomics., 16:318, 2015. doi: 10.1186/s12864-015-1514-4.
Tissue Antigens., 80(4):305-16, 2012. doi: 10.1111/j.1399-0039.2012.01941.x.
Hosomichi Group
Latest genome analysis technologies, such as next generation sequencer, measurements of biomolecules as single cell level, and bioinformatics for visualizing biological queries, reveal interaction network of disease causing neo-self antigen. Especially, integrated data analyses of genome, transcriptome and regulome sequencing have a big advantage for opening the black box of biological mechanisms to understand complex neo-self antigen diseases. Here, we would like to establish tools of integrated analyses of genome, gene expression, histone modification, DNA methylation and distal enhancer to understand immunological network centering on HLA molecular, named HLA-omics. The definition of the immunological mechanisms for neo-self antigen can open up doors of disease control and prevention. In the future, extending the knowledge of neo-self mechanisms will have the potential to become new medical treatment as big benefits for our health.
PLoS Genet., 12(4):e1005893, 2016. doi: 10.1371/journal.pgen.1005893.
Sci Rep., 6:20611, 2016. doi: 10.1038/srep20611.
J Hum Genet., 60(11):665-73, 2015. doi: 10.1038/jhg.2015.102.
BMC Genomics., 15:645, 2014. doi: 10.1186/1471-2164-15-645.
BMC Genomics. , 14:355, 2013. doi: 10.1186/1471-2164-14-355.
